An effective resonance integral.
More...
#include "pinspec/process.py"
|
|
| _name |
| | The name of the effective resonance integral.
|
| |
|
| _RI |
| | The DERIVED tally type for the effective resonance integral data.
|
| |
|
| _flux |
| | The pinspec.FLUX tally used to compute the effective resonance integral.
|
| |
|
| _rate |
| | The REACTION_RATE tally used to compute the effective resonance integral.
|
| |
|
| _num_RIs |
| | The number of resonance integral energy bands.
|
| |
An effective resonance integral.
This class represents an effective resonance integral computed by a PINSPEC monte carlo simulation. Two tallies - one reaction rate and one flux - are required to form a RIEff class object.
| def pinspec.process.RIEff.__init__ |
( |
|
self, |
|
|
|
tally1, |
|
|
|
tally2, |
|
|
|
name = '' |
|
) |
| |
RIEff constructor.
- Parameters
-
| self | the RIEff object pointer |
| tally1 | one of the tallies needed to compute an  |
| tally2 | one of the tallies needed to compute an 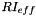 |
| name | an optional string for the name of the resonance integral |
| def pinspec.process.RIEff.computeRIs |
( |
|
self, |
|
|
|
tally1, |
|
|
|
tally2 |
|
) |
| |
Computes the resonance integrals from the two tallies.
This method checks that one of the tallies input is a reaction rate and the other is a flux, with the same bin edges. If the batch statistics for both tallies have been computed, it computes the resonance integral as follows:
2 log_array = numpy.log(edges[1:] / edges[0:edges.size-1])
3 RI_eff = (reaction_rate / flux).multiplyDoubles(log_array)
- Parameters
-
| self | the RIEff object pointer |
| tally1 | one of the tallies needed to compute an 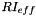 |
| tally2 | one of the tallies needed to compute an 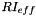 |
| def pinspec.process.RIEff.getEnergyBands |
( |
|
self | ) |
|
Returns an array of the resonance integral energy bands.
The energy bands are the tally bin edges for the reaction rate and flux tally forming this effective resonance integral.
- Parameters
-
| self | the RIEff object pointer |
- Returns
- a numpy array of the resonance integral energy bands (eV)
| def pinspec.process.RIEff.getEnergyBandsCenters |
( |
|
self | ) |
|
Returns an array of the resonance integral energy band centers for each tally bin.
- Parameters
-
| self | the RIEff object pointer |
- Returns
- a numpy array of the resonance integral energy band centers
| def pinspec.process.RIEff.getIntegrals |
( |
|
self | ) |
|
Returns an array of the resonance integral batch averaged values for each tally bin.
- Parameters
-
| self | the RIEff object pointer |
- Returns
- a numpy array of the resonance integrals
| def pinspec.process.RIEff.getName |
( |
|
self | ) |
|
Returns the name of the RIEff object.
Returns an empty string if no name has been specified by the user.
- Parameters
-
| self | the RIEff object pointer |
- Returns
- a string with the name of the RIEff
| def pinspec.process.RIEff.getNumIntegrals |
( |
|
self | ) |
|
Returns the number of resonance integral energy bands.
The number of integrals is equivalent to the number of tally bins for the reaction rate and flux tallies.
- Parameters
-
| self | the RIEff object pointer |
- Returns
- the number of resonance integrals
| def pinspec.process.RIEff.getRelativeError |
( |
|
self | ) |
|
Returns an array of the resonance integral batch relative errors for each tally bin.
- Parameters
-
| self | the RIEff object pointer |
- Returns
- a numpy array of the resonance integral relative errors
| def pinspec.process.RIEff.getRITally |
( |
|
self | ) |
|
Returns a reference to this RIEff object.
- Parameters
-
| self | RIEff the object pointer |
- Returns
- a reference to the RIEff object
| def pinspec.process.RIEff.getStandardDeviation |
( |
|
self | ) |
|
Returns an array of the resonance integral batch standard deviations for each tally bin.
- Parameters
-
| self | the RIEff object pointer |
- Returns
- a numpy array of the resonance integral standard deviations
| def pinspec.process.RIEff.getVariances |
( |
|
self | ) |
|
Returns an array of the resonance integral batch variances for each tally bin.
- Parameters
-
| self | the RIEff object pointer |
- Returns
- a numpy array of the resonance integral variances
| def pinspec.process.RIEff.outputRItoFile |
( |
|
self, |
|
|
|
filename = '' |
|
) |
| |
Prints the resonance integral data and batch statistics to a file.
Since the effective resonance integral is stored as a DERIVED tally type, this method prints the resonance integral data to a file using the Tally::outputBatchStatistics() method. An auto-generated filename will be created with the format 'tally-#.data' where # is an auto-incremented integer for each tally output data file created.
- Parameters
-
| self | the RIEff object pointer |
| filename | An optional filename for the output file |
| def pinspec.process.RIEff.printRI |
( |
|
self, |
|
|
|
uncertainties = False |
|
) |
| |
Prints a formatted table of the effective resonance integrals to the screen.
The resonance integrals and their uncertainties (optional) will be printed as a formatted table to the screen.
- Parameters
-
| self | the RIEff object pointer |
| uncertainties | whether or not to print tally statistics (default is false) |
- Returns
- a reference to the RIEff object
| def pinspec.process.RIEff.setName |
( |
|
self, |
|
|
|
name = '' |
|
) |
| |
Sets the name of this effective resonance integral.
This is useful when one wishes to print the resonance integral values to the screen or a file since it will be identifiable by the user-defined name.
- Parameters
-
| self | the RIEff object pointer |
| name | the name of the RIEff object |
The documentation for this class was generated from the following file: